Automating with Github Actions
I learned about Github Actions at a NICAR workshop in the spring but it was a recent Knight Center for Journalism in the Americas course where they were covered along with an introduction to Amazon Web Service S3 storage that provided a nudge to explore both in a project.
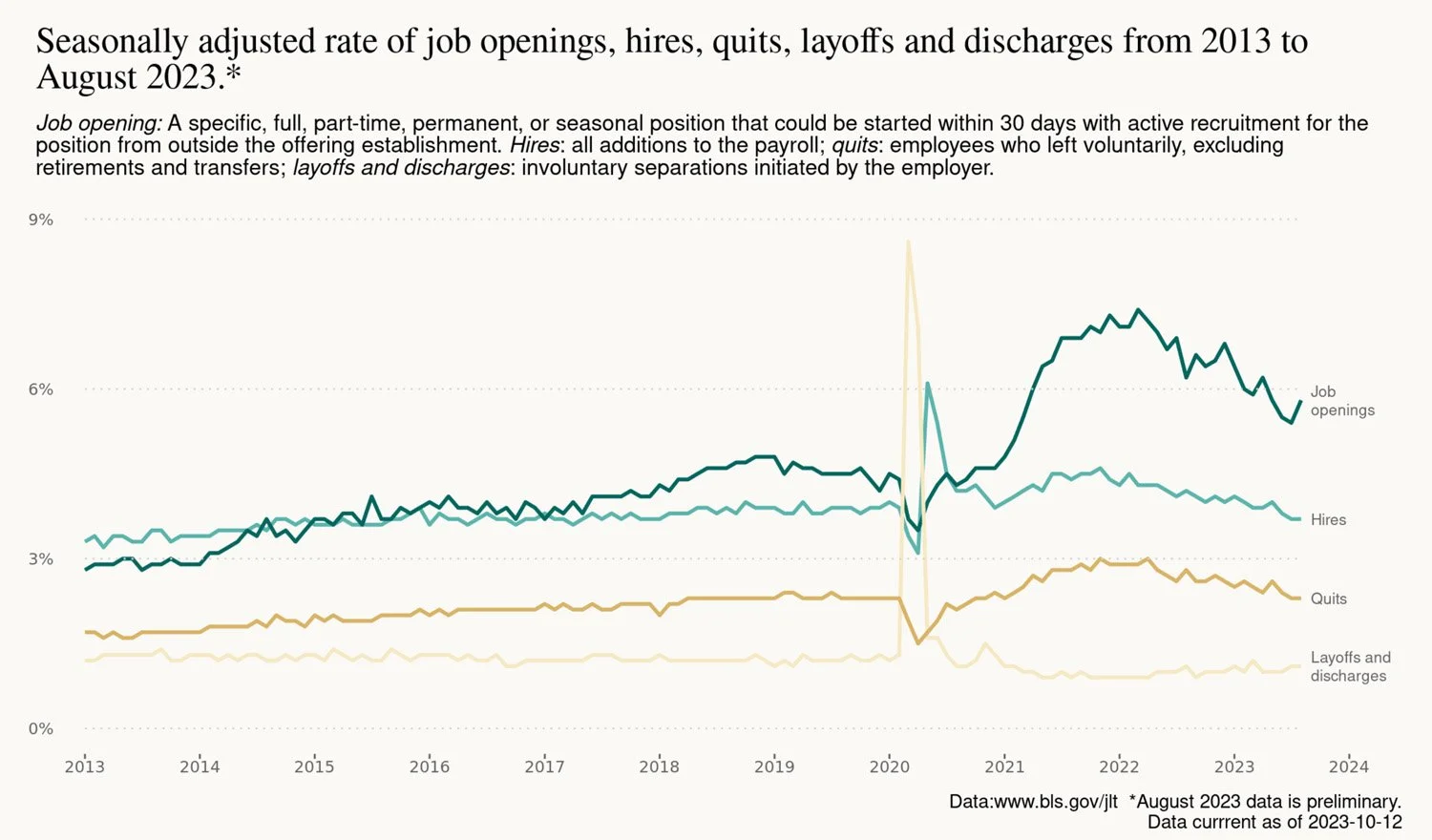
As a test case, I started with some data from the Job Openings and Labor Turnover Survey, commonly referred to as JOLTS, published by the Bureau of Labor Statistics. After sorting out AWS permissions and wrangling some data, I sent a csv file to an AWS S3 bucket to serve as a benchmark future scripts check to determine if data on the BLS API is old or new. (If I weren’t interested in exploring AWS, the same result could be accomplished within the Github repository.)
The data in the report and all the charts, tables and text are updated automatically using Github Actions when new data is released by the Bureau of Labor Statistics.
With the benchmark file set and permissions established for who can read and write to the AWS bucket, I wrote an R script to compare the data being offered on the BLS API with the last known JOLTS release stored in the AWS S3 bucket. If they are the same, no new data has been released and nothing else happens. If the files are different, the script writes the new data as a csv file to the Github repository, and sends a copy to the AWS S3 bucket to serve as the new benchmark.
If I were running this report manually on my own computer, the language in the above script would be used in an R Markdown script that can output the analysis as a document, html page or pdf. It would include additional code to generate tables, text and visualizations detailing changes in the data. Since I want to remove myself from the process, those steps will be parceled out to multiple scripts that run when initiated by specific events using Github Actions. The end result will be the same, though.
Github Actions uses a set of instructions in a yaml file to perform work. The yaml file specifies the type of virtual computer to create, what software and packages to load, and any actions or commands you want the virtual machine to perform. In this case, the yaml file tells Github Actions to install R on a Linux machine, load the packages required by the R script, copy the project’s Github repository onto the virtual machine, run the R script and send any output back to the Github repository.
To automate the project, a yaml file needs to know when it should run. In this case, the BLS publishes JOLTS data every few weeks at 10 a.m. Eastern Time, though the interval between releases and the day the data is released varies. This year the data has been released on Tuesdays, Wednesdays and Thursdays so I set the yaml script to run at 10:10 a.m. Monday through Friday just in case they feel rebellious. In addition to telling the virtual computer when to run, the yaml file tells it to execute the R script that checks for new data. This script will lead a dull life, running fruitlessly most days except for the rare instance every few weeks when new data is detected.
On those exciting days a different yaml file, configured to run when the new csv file is saved to the Github repository, springs to action. This yaml file contains instructions to run the R Markdown script that generates the report detailing changes in the JOLTS data. Once the report is written, the yaml instructions start a third R script that sends a dated copy of the report to the AWS S3 bucket for archiving. The yaml file’s final instruction is to copy the new report file from the virtual machine to the Github repository, enabling Github pages to publish the report.
Github Actions communicates when your runs fail; sorting out why takes more investigation.
I will get an email each time an action runs. Most days, it will just tell me no new data is available. Once every few weeks, though, I’ll get an email that the new report has been generated and published so I can review it for errors. It took more than a few mistakes to get right, but both AWS S3 and Github Actions are going to be a useful tools moving forward. A few tips and helpful resources that helped me get over the inevitable bumps include:
· Sorting out AWS S3 permissions, specifically limiting read and write privileges: https://www.gormanalysis.com/blog/connecting-to-aws-s3-with-r/
· Getting to know the aws.s3 R package: https://blog.djnavarro.net/posts/2022-03-17_using-aws-s3-in-r/
· Read the docs: Policies and Permissions in Amazon S3: https://docs.aws.amazon.com/AmazonS3/latest/userguide/access-policy-language-overview.html
· Read the docs Part II: Triggering a workflow (the ephemeral GITHUB_TOKEN will not trigger a push event workflow): https://docs.github.com/en/actions/using-workflows/triggering-a-workflow
· Ordinarily, AWS credentials get stored in the .Renviron file locally. For Github Actions, they need to be stored as a secret, then written to the .Renviron file of the virtual machine so they can be accessed by scripts. A simple solution: https://stackoverflow.com/questions/65926899/how-can-i-get-an-r-environment-via-sys-getenv-with-github-actions-using-secret
· Although R scripts executed normally on the virtual machines, the R Markdown required a different file path when reading in a file from the repository. Using ../folder_name/file_name did the trick. Determining the error was related to the file path and not an environmental variable, permission or extra yaml space somewhere took a while.
· www.yamllint.com will test for some of the yaml syntax errors like extra spaces that trip up Github Actions so you can use commits for other changes.